Hello Learners…
Welcome to the blog…
Table Of Contents
- Introduction
- Meditron An Open Source Medical Large Language Models (LLMs)
- Meditron Model Details
- Packages Requirements For Meditron
- How to use Meditron?
- Summary
- References
Introduction
In this post, we discuss about new large language model Meditron An Open Source Medical Large Language Models (LLMs).
MEDITRON builds on Llama-2 (through our adaptation of Nvidia’s Megatron-LM distributed trainer), and extends pretraining on a comprehensively curated medical corpus, including selected PubMed articles, abstracts, and internationally-recognized medical guidelines
Meditron An Open Source Medical Large Language Models (LLMs)
Large language models (LLMs) can potentially democratize access to medical knowledge.
While many efforts have been made to harness and improve LLMs’ medical knowledge and reasoning capacities, the resulting models are either closed-source (e.g., PaLM, GPT-4) or limited in scale (<= 13B parameters), which restricts their abilities.
In this work, They improve access to large-scale medical LLMs by releasing MEDITRON: a suite of open-source LLMs with 7B and 70B parameters adapted to the medical domain
GitHub URL:
Research Paper URL:
Meditron Model Details
- Developed by: EPFL LLM Team
- Model type: Causal decoder-only transformer language model
- Language(s): English (mainly)
- Model License: LLAMA 2 COMMUNITY LICENSE AGREEMENT
- Code License: APACHE 2.0 LICENSE
- Continue-pretrained from model: Llama-2-70B
- Context length: 4k tokens
- Input: Text only data
- Output: Model generates text only
- Status: This is a static model trained on an offline dataset. Future versions of the tuned models will be released as we enhance model’s performance.
- Knowledge Cutoff: August 2023
- Trainer: epflLLM/Megatron-LLM
- Paper: Meditron-70B: Scaling Medical Pretraining for Large Language Models
Packages Requirements For Meditron
To run this model, we have to install the necessary packages:
vllm >= 0.2.1 transformers >= 4.34.0 datasets >= 2.14.6 torch >= 2.0.1
How to use Meditron?
We can load Meditron model directly from the HuggingFace model hub as follows:
from transformers import AutoTokenizer, AutoModelForCausalLM
tokenizer = AutoTokenizer.from_pretrained("epfl-llm/meditron-70B")
model = AutoModelForCausalLM.from_pretrained("epfl-llm/meditron-70B")Medical Training Data For Meditron
They release code to download and pre-process the data used to train Meditron.
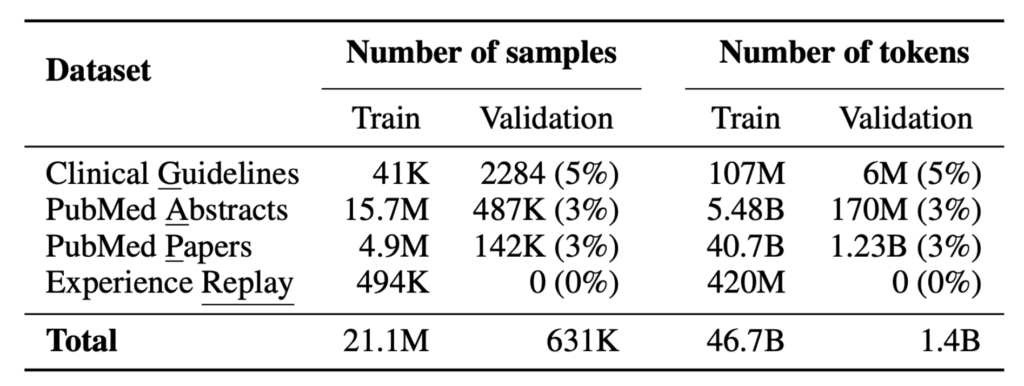
MediTron’s domain-adaptive pre-training corpus GAP-Replay combines 48.1B tokens from four corpora:
- Clinical Guidelines: a new corpus of 46K clinical practice guidelines from various healthcare-related sources, including hospitals and international organizations,
- Paper Abstracts: 16.1M abstracts extracted from closed-access PubMed and PubMed Central papers,
- Medical Papers: full-text articles extracted from 5M publicly available PubMed and PubMed Central papers.
- Replay dataset: 400M tokens of general domain pretraining data sampled from RedPajama-v1.
Uses Of Meditron
Meditron-70B is being made available for further testing and assessment as an AI assistant to enhance clinical decision-making and democratize access to an LLM for healthcare use. Potential use cases may include but are not limited to:
- Medical exam question answering
- Supporting differential diagnosis
- Disease information (symptoms, cause, treatment) query
- General health information query
It is possible to use this model to generate text, which is useful for experimentation and understanding its capabilities. It should not be used directly for production or work that may impact people.
They do not recommend using this model for natural language generation in a production environment, finetuned or otherwise.
MEDITRON achieves a 6% absolute performance gain over the best public baseline in its parameter class and 3% over the strongest baseline from Llama-2. Compared to closed-source LLMs, MEDITRON-70B outperforms GPT-3.5 and Med-PaLM and is within 5% of GPT-4 and 10% of Med-PaLM-2
Summary
The blog emphasizes the importance of open-source development and provides access to the code for corpus curation and the MEDITRON model weights.
